Hanyang University
Bioinformatics, Proteomics
NovoRank
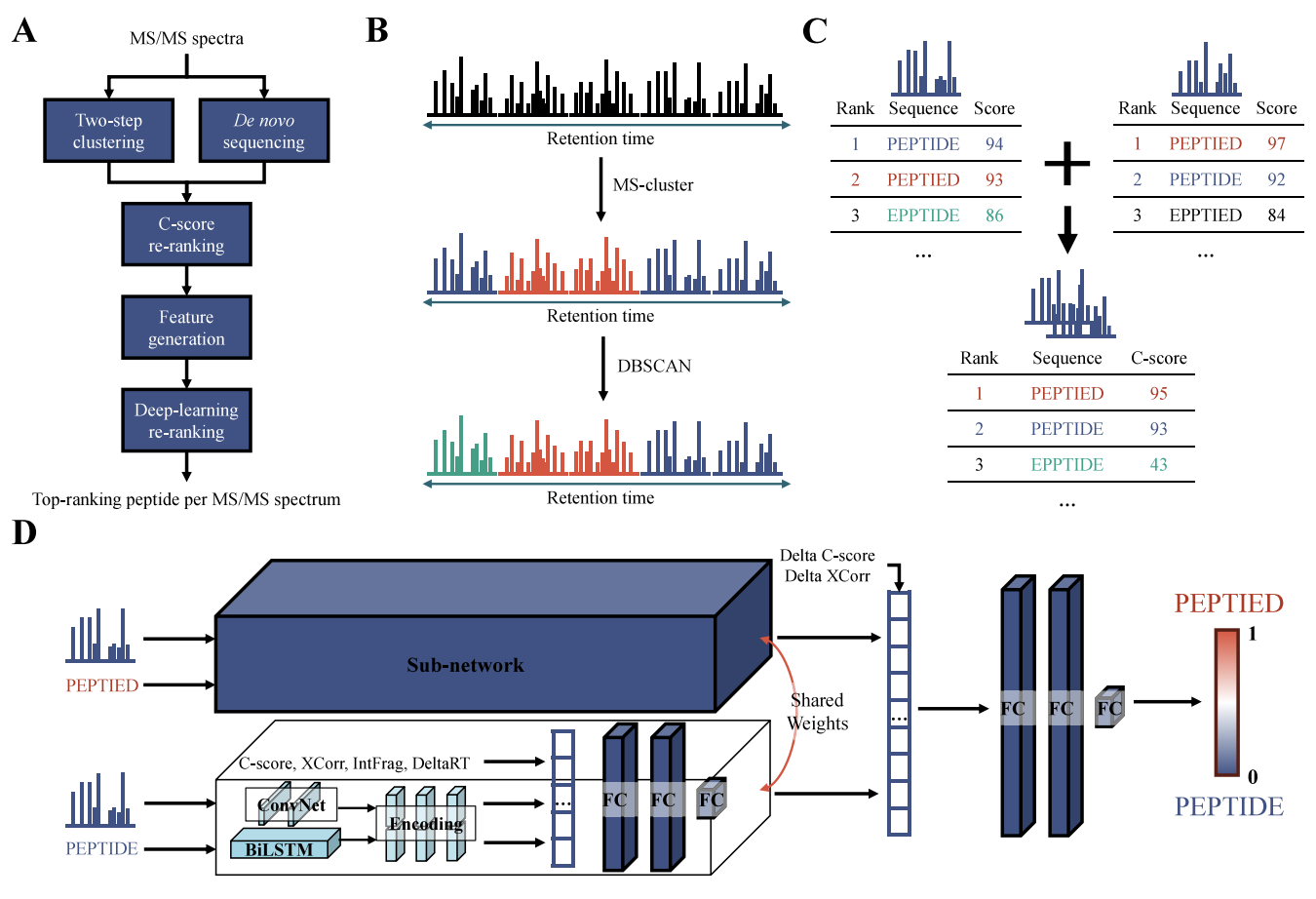
NovoRank is a machine learning-based post-processing tool designed to improve peptide identification accuracy.
Summary
Overview
De novo sequencing is a crucial technique in proteomics that identifies peptide sequences directly from experimental data, without relying on existing protein databases. This method plays a key role in identifying novel peptides, such as neoantigen discovery. However, conventional de novo sequencing tools consider only individual spectrum and rely solely on imperfect scoring functions, often leading to erroneous peptide identifications. NovoRank enhances the reliability and accuracy of de novo sequencing by re-ranking candidate sequences using the comprehensive analysis of similar spectra, assuming that they originate from the same peptide species.
Keywords
- Clustering
- Deep Learning
- Data Analytics
Dataset
The main dataset used in NovoRank consists of MS/MS spectra (.mgf), obtained from mass spectrometry experiments and containing peptide fragmentation information.
For more details, please refer to the link at the top of this section and here.
Contribution
- Introduced Two-Step Clustering Method: To effectively classify similar spectra, Spectral Clustering was applied for initial grouping, followed by DBSCAN to perform fine-grained clustering within the first-stage clusters, ensuring that similar spectra are more precisely grouped together.
- Proposed C-Score: A new scoring method (C-Score) was introduced to identify more reliable candidate peptides within clusters. The C-Score is calculated by normalizing the sum of each peptide's de novo scores by the cluster size. This method assigns higher scores to peptides with higher de novo scores and those that appear more frequently within the cluster, allowing for a fair comparison between clusters of different sizes. This approach ensures that peptides with a higher likelihood of being the correct sequence are ranked higher, leading to more accurate results.
- Applied Deep Learning Model: Developed a multi-modal deep learning model that simultaneously takes spectrum, sequence, and tabular data as inputs to assign the best peptide. By selecting the most desirable peptide from the top two candidates, the model improved identification accuracy.
- Applicable to Existing De Novo Sequencing Tools: Applied to various de novo peptide sequencing tools, improving peptide identification performance by enhancing the precision and recall.
Results
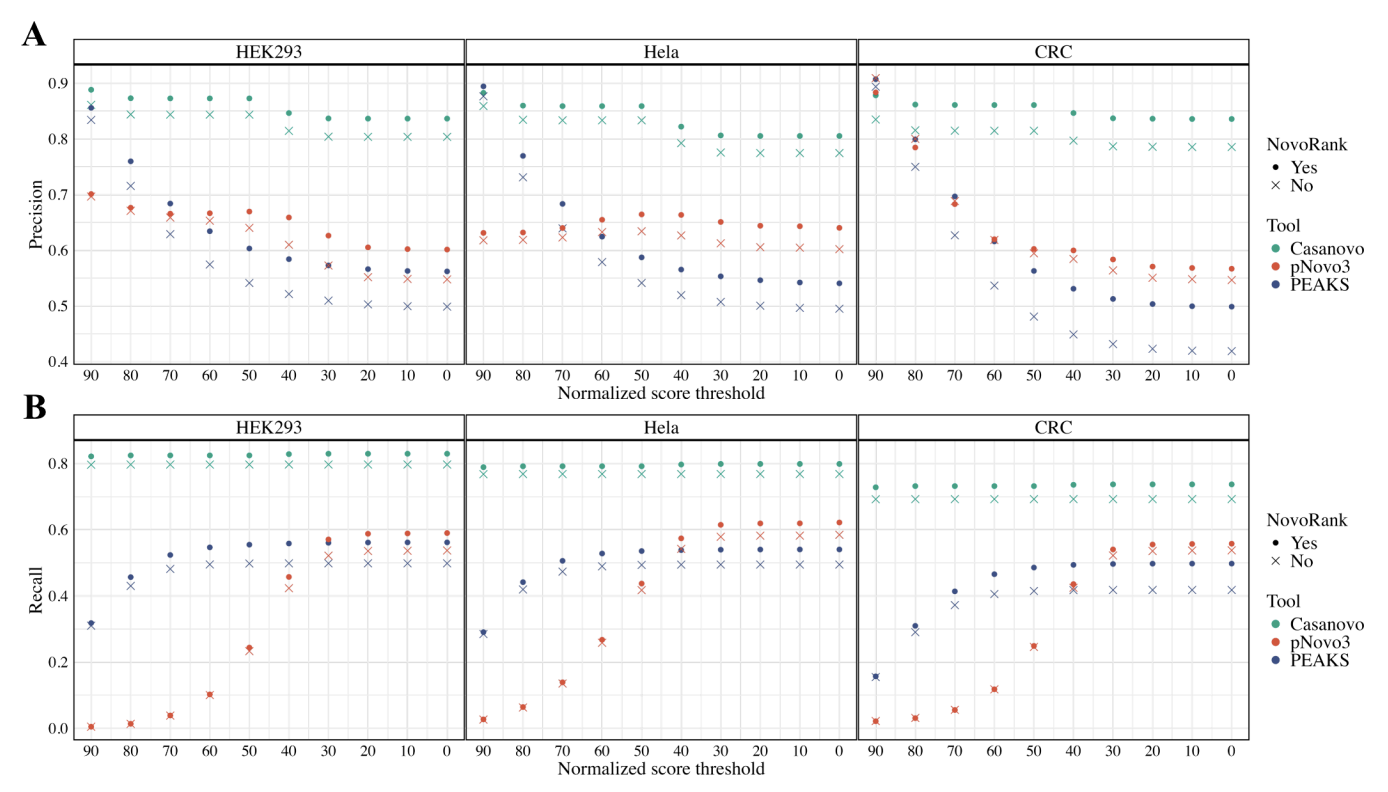
- Precision and recall increased by an average of 4.6% and 4.5%, with improvements of 3.8% and 3.6% specifically in Casanovo, a state-of-the-art (SOTA) model.
- Casanovo, pNovo3, and PEAKS achieved an increase of 0.37%−0.61%, 7.52%−18.80%, and 3.06%−4.24% in correct peptide identifications, respectively. Although some peptides were missed, the number of newly identified peptides was greater.
※ De novo sequencing tools
1. PEAKS: Algorithm-based commercial tool
2. pNovo3: Tool combining algorithm and machine learning
3. Casanovo: Deep learning-based tool using transformer (SOTA)